Help
Abbreviations
- SNV:
- single nucleotide variant
- nt:
- nucleotide
- bp:
- base pair
- indel:
- insertion or deletion
- ref:
- reference
- alt:
- alternate
Notes:
- 1 bp insertion probability = proportion of indel outcomes that are 1 bp insertions
- 1 bp frameshift frequency = proportion of indel outcomes that induce a 1 bp frameshift
- 2 bp frameshift frequency = proportion of indel outcomes that induce a 2 bp frameshift
- (Overall) frameshift frequency = proportion of indel outcomes that induce a frameshift (1
or 2 bp)
Visualizations
Frameshift, variant load, variant impact on frameshift
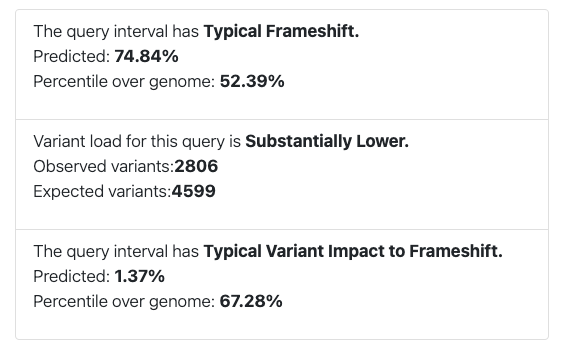
Frameshift predictions
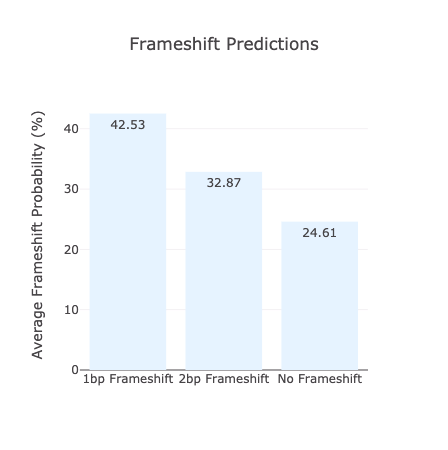
Nucleotide Substitution Frequency Map
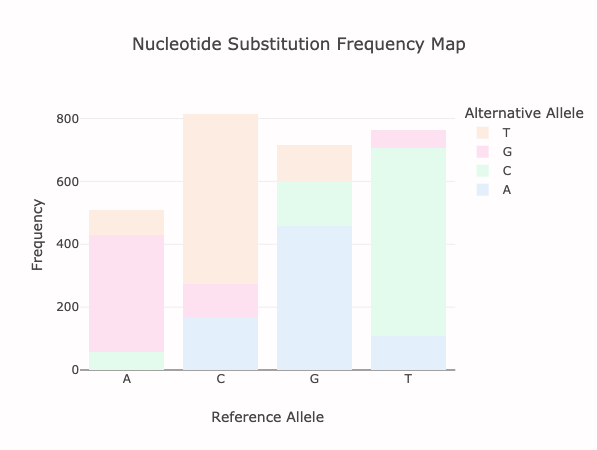
Histograms for alternate 1 bp insertion, 1 bp frameshift, 2 bp frameshift
Table
| # | Column header | Description |
|---|---|---|
| 1 | Variant ID | unique identifier for the SNV |
| 2 | Variant Position | the genomic position of the SNV |
| 3 | Reference Allele | the nt present in the reference genome |
| 4 | Alternate Allele | the nt present in an alternate genome |
| 5 | PAM ID | unique identifier for the PAM site PAM ID is defined as [`genename`] | [`num`] where `num` identifies the PAM site of a specific gene. The `num` of the PAM site increases as the position of the first nt of the 60 bp section of the gene-of-interest increases on the positive strand. |
| 6 | PAM Range | describes the position of the 60 bp sequence CROTON uses to make
predictions
PAM Range is defined as [`strand`] : [`start`] – [`end`] where `strand` indicates if the PAM site is sense (+) or antisense (–), and `start` and `end` indicate the position of the first and last nt, respectively, of the CROTON input sequence. |
| 7 | Max Variant Effect | The maximum absolute difference between CROTON’s set of predictions for the reference and alternate sequence inputs. [This is the maximum between CROTON’s reference and alternate predictions for the set of statistics: 1 bp insertion probability, 1 bp frameshift frequency, and 2 bp frameshift frequency (shown in table), as well as deletion frequency, 1 bp deletion frequency, and overall frameshift frequency (not shown in table).] |
| 8 | Ref. 1 bp insertion | CROTON’s 1 bp insertion probability prediction for the reference sequence |
| 9 | Ref. 1 bp frameshift | CROTON’s 1 bp frameshift frequency prediction for the reference sequence |
| 10 | Ref. 2 bp frameshift | CROTON’s 2 bp frameshift frequency prediction for the reference sequence |
| 11 | Alt. 1 bp insertion | CROTON’s 1 bp insertion probability prediction for the alternate sequence |
| 12 | Alt. 1 bp frameshift | CROTON’s 1 bp frameshift frequency prediction for the alternate sequence |
| 13 | Alt. 2 bp frameshift | CROTON’s 2 bp frameshift frequency prediction for the alternate sequence |
| 14 | Ref_seq and alt_seq | The 60 bp PAM-containing sequence in the reference and alternate genome (the 0-index PAM site is at position 33). There is a single nucleotide difference between the reference and alternate sequences. |